Viewing the Human Brain Through Noninvasive Tools
- Reviewed9 Mar 2023
- Author Susan Rojahn
- Source BrainFacts/SfN
Scientists add new techniques and technologies to their repertoire as they emerge, helping to deepen our understanding of the brain and suggest new ways to assist people whose lives are affected by brain disorders and diseases. And while many of the methods and tools used in neuroscience research are too invasive to use in humans, several methods for imaging human brain function do not require holes in the skull or other lasting physical changes.
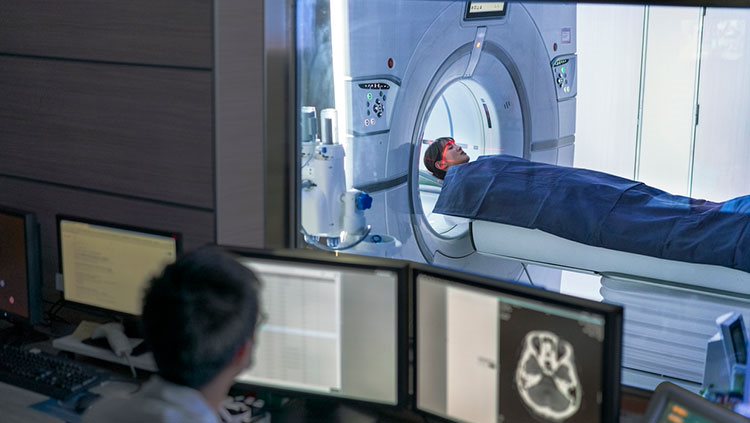
Functional MRI (fMRI) can be used to follow changes in the brain activity of a person lying inside an MRI scanner. The machine is tuned so that it detects blood flow as well as differences in oxygen-rich and oxygen-poor blood, based on the idea that more active regions of the brain need more oxygen and nutrients, which are supplied by fresh oxygenated blood. While this is an indirect indication of neuron activity, it can pinpoint brain activity to fairly small regions.
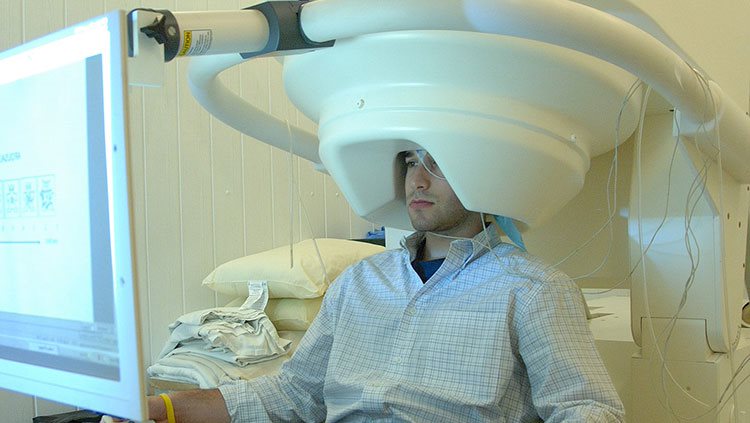
fMRI provides an indirect view of neuron activity, but magnetoencephalography (MEG) detects actual electrical currents coursing through groups of neurons. When neuron activities are synchronized, their combined electrical currents produce weak magnetic fields that MEG equipment can detect. A person undergoing the procedure sits or lies down, with his or her head surrounded by a helmet-shaped device that can sense magnetic fields. MEG has been useful in a variety of studies: from how the auditory cortex responds to sounds to identifying where epileptic seizures start in a patient’s brain. MEG is useful for detecting rapid changes in brain activity (temporal resolution), but it does not provide the precise location of that activity (spatial resolution). For this reason, researchers can combine MEG data with fMRI data to obtain good anatomical detail from fMRI and high-speed readings of brain activity from MEG.
Near-infrared spectroscopy (NIRS) is similar to fMRI in that it monitors the flow of oxygenated blood as a way to estimate neuron activity. A major difference is that NIRS is only useful for measuring activity near the surface of the brain and does not provide as much detail; however, it is far less expensive and cumbersome than fMRI. NIRS is also more comfortable for the person undergoing the procedure, as the setup essentially involves wearing a cap with wiring hooked to it. Some of the wires transmit harmless laser beams (~1 megawatt of power or less) into the brain while others detect the light after it travels through the brain. NIRS can be used to determine the extent of brain injuries and to monitor oxygen levels in the brains of patients under anesthesia. Because of its portability, NIRS is very useful for studying brain activity during tasks — such as driving down the highway — that can’t take place inside an fMRI scanner.
Positron emission tomography (PET) detects short-lived radioactive compounds that have been injected into the bloodstream. The radioactive compounds could be oxygen or glucose, or they might be a neurotransmitter. PET traces where these compounds go in the body. The location of labeled oxygen can indicate blood flow, while a labeled neurotransmitter can show which brain regions are using that signaling molecule. PET can also detect the amyloid plaques that are associated with Alzheimer’s disease; this technique could one day enable us to identify the disease in its early stages. Although PET has good temporal resolution like MEG, it lacks the detailed spatial resolution of MRI.
Some methods used in human research can change brain activity. In transcranial magnetic stimulation (TMS), a coil that generates a magnetic field is placed near a person’s head. The magnetic field can penetrate the skull, temporarily activating or silencing a region of the cortex. TMS is used to treat psychiatric disorders such as anxiety, depression, and post-traumatic stress disorder and could be an effective option for patients with conditions that do not respond to medications.
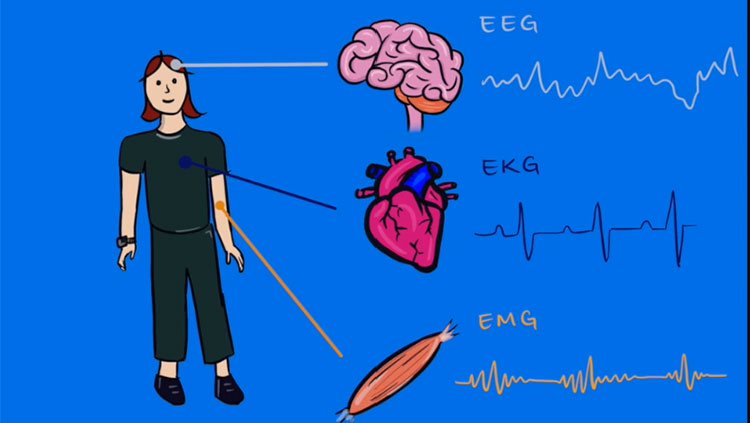
Although neuroscience has progressed by leaps and bounds since neurons were first viewed under a microscope, many phenomena observed with these techniques are not fully understood. For example, mysteries still surround data obtained with electroencephalography (EEG). EEG shows that several different brain regions have characteristic rhythms or oscillations — one pattern in the visual cortex, another in the sensory motor cortex, and so on. Even though this method of examining brain activity has been used since 1929, the generation of these patterns (sometimes called brain waves) at the level of neural circuits is not well understood.
One branch of neuroscience that can help bridge findings from the microscopic to the whole-brain level is computational neuroscience. Researchers in this field develop theories or models about how the brain processes information, then test these models against real-world data. For example, they can examine the data and images from EEG or fMRI, then develop mathematical models to explain the underlying neuron and circuit activity. Data from many methods — electrophysiology, molecular studies, anatomy, and functional brain scans — can all contribute to these computational models.
Adapted from the 8th edition of Brain Facts by Susan Rojahn.
CONTENT PROVIDED BY
BrainFacts/SfN
References
Bao, W., Jia, H., Finnema, S., Cai, Z., Carson, R. E., & Huang, Y. H. (2017). PET Imaging for Early Detection of Alzheimer's Disease: From Pathologic to Physiologic Biomarkers. PET clinics, 12(3), 329–350. https://doi.org/10.1016/j.cpet.2017.03.001
Berman, M. G., Jonides, J., & Nee, D. E. (2006). Studying Mind and Brain with fMRI. Social cognitive and affective neuroscience, 1(2), 158–161. https://doi.org/10.1093/scan/nsl019
Bögershausen, N., & Wollnik, B. (2013). Unmasking Kabuki syndrome. Clinical genetics, 83(3), 201–211. https://doi.org/10.1111/cge.12051
Boyden E. S. (2015). Optogenetics and the Future of Neuroscience. Nature neuroscience, 18(9), 1200–1201. https://doi.org/10.1038/nn.4094
Caraci, F., Leggio, G. M., Salomone, S., & Drago, F. (2017). New Drugs in Psychiatry: Focus on New Pharmacological Targets. F1000Research, 6, 397. https://doi.org/10.12688/f1000research.10233.1
Carter M. and Shieh J. C. (2015). Guide to Research Techniques in Neuroscience. Academic Press. p 164.
Carter N. P. (2007). Methods and Strategies for Analyzing Copy Number Variation Using DNA Microarrays. Nature genetics, 39(7 Suppl), S16–S21. https://doi.org/10.1038/ng2028
Chefer, V. I., Thompson, A. C., Zapata, A., & Shippenberg, T. S. (2009). Overview of Brain Microdialysis. Current protocols in neuroscience, Chapter 7, Unit 7.1. https://doi.org/10.1002/0471142301.ns0701s47
Clancy, S. (2008). Copy Number Variation. Nature Education, 1(1):95. https://www.nature.com/scitable/topicpage/copy-number-variation-445/
Cohen M. X. (2017). Where Does EEG Come From and What Does It Mean?. Trends in neurosciences, 40(4), 208–218. https://doi.org/10.1016/j.tins.2017.02.004
Courtney, K. E., & Ray, L. A. (2014). Methamphetamine: An Update on Epidemiology, Pharmacology, Clinical Phenomenology, and Treatment Literature. Drug and alcohol dependence, 143, 11–21. https://doi.org/10.1016/j.drugalcdep.2014.08.003
Cui, X., Bray, S., Bryant, D. M., Glover, G. H., & Reiss, A. L. (2011). A Quantitative Comparison of NIRS and fMRI Across Multiple Cognitive Tasks. NeuroImage, 54(4), 2808–2821. https://doi.org/10.1016/j.neuroimage.2010.10.069
Flagel, S. B., Chaudhury, S., Waselus, M., Kelly, R., Sewani, S., Clinton, S. M., Thompson, R. C., Watson, S. J., Jr, & Akil, H. (2016). Genetic Background and Epigenetic Modifications in the Core of the Nucleus Accumbens Predict Addiction-like Behavior in a Rat Model. Proceedings of the National Academy of Sciences of the United States of America, 113(20), E2861–E2870. https://doi.org/10.1073/pnas.1520491113
Gratten, J., Wray, N. R., Keller, M. C., & Visscher, P. M. (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature neuroscience, 17(6), 782–790. https://doi.org/10.1038/nn.3708
Hämäläinen, M., Hari, R., Ilmoniemi, R. J., Knuutila, J., & Lounasmaa, O. V. (1993). Magnetoencephalography—Theory, Instrumentation, and Applications to Noninvasive Studies of the Working Human Brain. Reviews of modern Physics, 65(2), 413. https://journals.aps.org/rmp/abstract/10.1103/RevModPhys.65.413
Hanrieder, J., Phan, N. T., Kurczy, M. E., & Ewing, A. G. (2013). Imaging Mass Spectrometry in Neuroscience. ACS chemical neuroscience, 4(5), 666–679. https://doi.org/10.1021/cn400053c
Heather, J. M., & Chain, B. (2016). The Sequence of Sequencers: The History of Sequencing DNA. Genomics, 107(1), 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003
Heidenreich, M., & Zhang, F. (2016). Applications of CRISPR-Cas Systems in Neuroscience. Nature reviews. Neuroscience, 17(1), 36–44. https://doi.org/10.1038/nrn.2015.2
Herbst, S. M., Proepper, C. R., Geis, T., Borggraefe, I., Hahn, A., Debus, O., Haeussler, M., von Gersdorff, G., Kurlemann, G., Ensslen, M., Beaud, N., Budde, J., Gilbert, M., Heiming, R., Morgner, R., Philippi, H., Ross, S., Strobl-Wildemann, G., Muelleder, K., Vosschulte, P., … Hehr, U. (2016). LIS1-associated Classic Lissencephaly: A Retrospective, Multicenter Survey of the Epileptogenic Phenotype and Response to Antiepileptic Drugs. Brain & development, 38(4), 399–406. https://doi.org/10.1016/j.braindev.2015.10.001
Hopf, F. W., & Lesscher, H. M. (2014). Rodent Models for Compulsive Alcohol Intake. Alcohol (Fayetteville, N.Y.), 48(3), 253–264. https://doi.org/10.1016/j.alcohol.2014.03.001
Johnson, A. C., & Greenwood-Van Meerveld, B. (2016). The Pharmacology of Visceral Pain. Advances in pharmacology (San Diego, Calif.), 75, 273–301. https://doi.org/10.1016/bs.apha.2015.11.002
Kandel, E. R., Dudai, Y., & Mayford, M. R. (2014). The Molecular and Systems Biology of Memory. Cell, 157(1), 163–186. https://doi.org/10.1016/j.cell.2014.03.001
Lee, G. J., Park, J. H., & Park, H. K. (2008). Microdialysis Applications in Neuroscience. Neurological research, 30(7), 661–668. https://doi.org/10.1179/174313208X289570
Leroy, A., Foucher, J. R., Pins, D., Delmaire, C., Thomas, P., Roser, M. M., Lefebvre, S., Amad, A., Fovet, T., Jaafari, N., & Jardri, R. (2017). fMRI Capture of Auditory Hallucinations: Validation of the Two-Steps Method. Human brain mapping, 38(10), 4966–4979. https://doi.org/10.1002/hbm.23707
Liu, Z., Ding, L., & He, B. (2006). Integration of EEG/MEG with MRI and fMRI. IEEE engineering in medicine and biology magazine : the quarterly magazine of the Engineering in Medicine & Biology Society, 25(4), 46–53. https://doi.org/10.1109/memb.2006.1657787
Lodish H., Berk A., Zipurksy S. L. et. al., editors. (2000). Molecular Cell Biology, 4th edition. Freeman, p 94, 140, 147-148, 268-269.
Malik, A. N., Vierbuchen, T., Hemberg, M., Rubin, A. A., Ling, E., Couch, C. H., Stroud, H., Spiegel, I., Farh, K. K., Harmin, D. A., & Greenberg, M. E. (2014). Genome-wide Identification and Characterization of Functional Neuronal Activity-Dependent Enhancers. Nature neuroscience, 17(10), 1330–1339. https://doi.org/10.1038/nn.3808
Mayford, M., Siegelbaum, S. A., & Kandel, E. R. (2012). Synapses and Memory Storage. Cold Spring Harbor perspectives in biology, 4(6), a005751. https://doi.org/10.1101/cshperspect.a005751
Maze, I., Shen, L., Zhang, B., Garcia, B. A., Shao, N., Mitchell, A., Sun, H., Akbarian, S., Allis, C. D., & Nestler, E. J. (2014). Analytical Tools and Current Challenges in the Modern Era of Neuroepigenomics. Nature neuroscience, 17(11), 1476–1490. https://doi.org/10.1038/nn.3816
National Human Genome Research Institute. (July 2017). An Overview of the Human Genome Project. Accessed July 17, 2017 at https://www.genome.gov/12011238/an-overview-of-the-human-genome-project/
National Institute of Mental Health. (2017). Brain Stimulation Therapies. Accessed July 17, 2017 at https://www.nimh.nih.gov/health/topics/brain-stimulation-therapies/brain-stimulation-therapies.shtml
Olgiati, S., Quadri, M., & Bonifati, V. (2016). Genetics of Movement Disorders in the Next-Generation Sequencing Era. Movement disorders, 31(4), 458–470. https://doi.org/10.1002/mds.26521
Perry, R. H., Blessed, G., Perry, E. K., & Tomlinson, B. E. (1980). Histochemical Observations on Cholinesterase Activities in the Brains of Elderly Normal and Demented (Alzheimer-type) Patients. Age and ageing, 9(1), 9–16. https://doi.org/10.1093/ageing/9.1.9
Purves D, Augustine GJ, Fitzpatrick D, et al., editors. (2008). Neuroscience. 4th edition. Sinauer Associates, Inc. p 3-5, 16-17, 19-21, 25-27, 181-187, 465, 559, 673-674, 715-717.
Sejnowski, T. J., Koch, C., & Churchland, P. S. (1988). Computational Neuroscience. Science (New York, N.Y.), 241(4871), 1299–1306. https://doi.org/10.1126/science.3045969
Sokolowski M. B. (2001). Drosophila: Genetics Meets Behaviour. Nature reviews. Genetics, 2(11), 879–890. https://doi.org/10.1038/35098592
Svoboda, K., & Yasuda, R. (2006). Principles of Two-Photon Excitation Microscopy and its Applications to Neuroscience. Neuron, 50(6), 823–839. https://doi.org/10.1016/j.neuron.2006.05.019
Turek, F. W., Pinto, L. H., Vitaterna, M. H., Penev, P. D., Zee, P. C., & Takahashi, J. S. (1995). Pharmacological and Genetic Approaches for the Study of Circadian Rhythms in Mammals. Frontiers in neuroendocrinology, 16(3), 191–223. https://doi.org/10.1006/frne.1995.1007
US National Library of Medicine, National Institutes of Health. (2017). Genetics Home Reference – Huntington Disease. Accessed July 17, 2017 at https://ghr.nlm.nih.gov/condition/huntington-disease#genes
Usdin, K., & Kumari, D. (2015). Repeat-mediated Epigenetic Dysregulation of the FMR1 Gene in the Fragile X-related Disorders. Frontiers in genetics, 6, 192. https://doi.org/10.3389/fgene.2015.00192
Yoshino, K., Oka, N., Yamamoto, K., Takahashi, H., & Kato, T. (2013). Functional Brain Imaging Using Near-infrared Spectroscopy During Actual Driving on an Expressway. Frontiers in human neuroscience, 7, 882. https://doi.org/10.3389/fnhum.2013.00882
What to Read Next
Also In Tools & Techniques
Trending
Popular articles on BrainFacts.org