Mapping and Imaging Neural Anatomy
- Reviewed27 Feb 2023
- Author Susan Rojahn
- Source BrainFacts/SfN
Our brains aren’t so simple. But that hasn’t stopped us from trying to understand their complexity: Cell atlases in development are attempting to map the nearly 200 billion cells within the human brain.
Anatomy is the study of structure — most often, the structure of biological organisms. For the brain, anatomy tends to center around the structure of neurons, which are among the most complex and diverse cell types in our bodies. Scientists were first able to observe neurons in the late 19th century, thanks to histological techniques that start with a very thin slice of brain tissue to which scientists apply stains or other compounds that add contrast or color to specific structures. They then view the tissue with a light microscope, which passes visible light through the thin slice and lenses that make the structures look up to 1,000 times larger than they do with the naked eye.
Histology is the study of how cells form tissues. Histological techniques can reveal changes in the density of cell types or the presence of molecules that can suggest a particular disease. These techniques have helped illuminate the brain changes underlying some neurodegenerative disorders. For example, histological methods have shown that an enzyme that breaks down acetylcholine is associated with the brain plaques and tangles of Alzheimer’s disease. And in the brains of Parkinson’s disease patients, histology has revealed the death of neurons that normally control movements through dopamine signaling.
Long after light microscopes gave scientists their first glimpses of neurons, a debate bubbled in the scientific community: Are neurons individual cells or a mesh of physically interconnected cell bodies? Neurons are so densely packed that the answer wasn’t clear until the 1950s, after the development of a new technology called electron microscopy. Electron microscopes can produce useful detailed images of cellular structures magnified many 100,000s of times by directing a beam of electrons through very thin slices of tissue, then enlarging and focusing the image with electromagnetic lenses. With this technology, researchers were finally able to see that neurons are not physically continuous but, instead, are individual cells.
Although they are individual cells, neurons do act in networks, communicating across small gaps called synapses, where the axon terminal of one cell meets a dendrite or cell body of another cell. One method for mapping the signaling pathways within these networks involves injecting radioactive molecules or “tracers” into the cell body of a neuron. Researchers monitor the movement of radioactivity down the neuron’s axon, showing where that neuronal path leads. A similar technique involves tracers that can actually travel across synapses, from one neuron to the next. Scientists have used such tracers to map the complex pathways by which information travels from the eyes to the visual cortex.
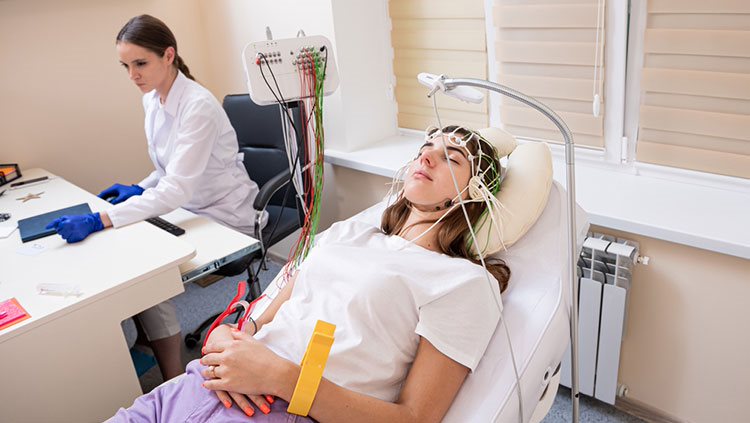
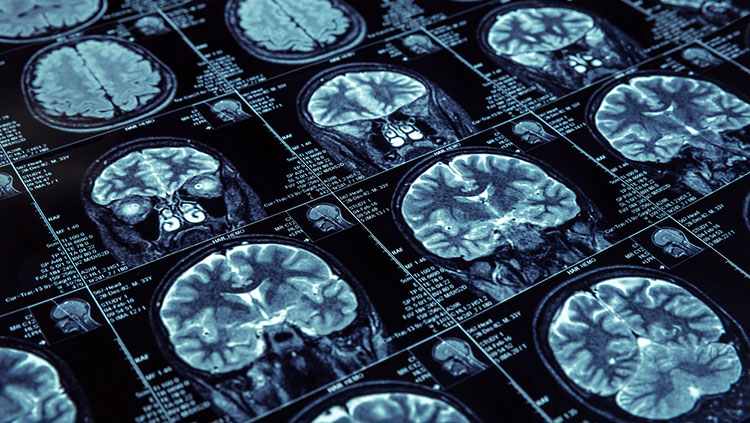
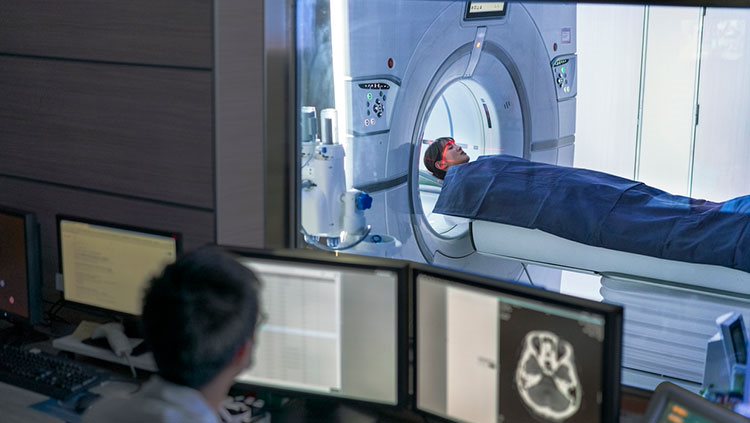
Another technique for examining brain anatomy is magnetic resonance imaging, or MRI. Developed in the 1980s, MRI is widely used by researchers and doctors to view a detailed image of brain structure. MRI equipment uses radio waves and strong magnets to create images of the brain based on the distribution of water within its tissues. MRI is harmless and painless to the person being scanned, although it does require sitting or lying in a narrow tube, and the procedure can be quite noisy. With an MRI scan, researchers can tell the difference between the brain’s gray matter and white matter. Gray matter consists of the cell bodies of neurons, as well as their dendrites and synapses. White matter mostly contains axons wrapped in the fatty myelin coating that gives these regions their white color. Based on the distribution of water in the tissues, MRI images clearly differentiate between cerebrospinal fluid, the water-rich cells of gray matter, and fatty white matter.
Adapted from the 8th edition of Brain Facts by Susan Rojahn.
CONTENT PROVIDED BY
BrainFacts/SfN
References
Berman, M. G., Jonides, J., & Nee, D. E. (2006). Studying Mind and Brain with fMRI. Social cognitive and affective neuroscience, 1(2), 158–161. https://doi.org/10.1093/scan/nsl019
Bögershausen, N., & Wollnik, B. (2013). Unmasking Kabuki syndrome. Clinical genetics, 83(3), 201–211. https://doi.org/10.1111/cge.12051
Boyden E. S. (2015). Optogenetics and the Future of Neuroscience. Nature neuroscience, 18(9), 1200–1201. https://doi.org/10.1038/nn.4094
Caraci, F., Leggio, G. M., Salomone, S., & Drago, F. (2017). New Drugs in Psychiatry: Focus on New Pharmacological Targets. F1000Research, 6, 397. https://doi.org/10.12688/f1000research.10233.1
Carter M. and Shieh J. C. (2015). Guide to Research Techniques in Neuroscience. Academic Press. p 164.
Carter N. P. (2007). Methods and Strategies for Analyzing Copy Number Variation Using DNA Microarrays. Nature genetics, 39(7 Suppl), S16–S21. https://doi.org/10.1038/ng2028
Chefer, V. I., Thompson, A. C., Zapata, A., & Shippenberg, T. S. (2009). Overview of Brain Microdialysis. Current protocols in neuroscience, Chapter 7, Unit 7.1. https://doi.org/10.1002/0471142301.ns0701s47
Clancy, S. (2008). Copy Number Variation. Nature Education, 1(1):95. https://www.nature.com/scitable/topicpage/copy-number-variation-445/
Cohen M. X. (2017). Where Does EEG Come From and What Does It Mean?. Trends in neurosciences, 40(4), 208–218. https://doi.org/10.1016/j.tins.2017.02.004
Courtney, K. E., & Ray, L. A. (2014). Methamphetamine: An Update on Epidemiology, Pharmacology, Clinical Phenomenology, and Treatment Literature. Drug and alcohol dependence, 143, 11–21. https://doi.org/10.1016/j.drugalcdep.2014.08.003
Cui, X., Bray, S., Bryant, D. M., Glover, G. H., & Reiss, A. L. (2011). A Quantitative Comparison of NIRS and fMRI Across Multiple Cognitive Tasks. NeuroImage, 54(4), 2808–2821. https://doi.org/10.1016/j.neuroimage.2010.10.069
Flagel, S. B., Chaudhury, S., Waselus, M., Kelly, R., Sewani, S., Clinton, S. M., Thompson, R. C., Watson, S. J., Jr, & Akil, H. (2016). Genetic Background and Epigenetic Modifications in the Core of the Nucleus Accumbens Predict Addiction-like Behavior in a Rat Model. Proceedings of the National Academy of Sciences of the United States of America, 113(20), E2861–E2870. https://doi.org/10.1073/pnas.1520491113
Gratten, J., Wray, N. R., Keller, M. C., & Visscher, P. M. (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature neuroscience, 17(6), 782–790. https://doi.org/10.1038/nn.3708
Hämäläinen, M., Hari, R., Ilmoniemi, R. J., Knuutila, J., & Lounasmaa, O. V. (1993). Magnetoencephalography—Theory, Instrumentation, and Applications to Noninvasive Studies of the Working Human Brain. Reviews of modern Physics, 65(2), 413. https://journals.aps.org/rmp/abstract/10.1103/RevModPhys.65.413
Hanrieder, J., Phan, N. T., Kurczy, M. E., & Ewing, A. G. (2013). Imaging Mass Spectrometry in Neuroscience. ACS chemical neuroscience, 4(5), 666–679. https://doi.org/10.1021/cn400053c
Heather, J. M., & Chain, B. (2016). The Sequence of Sequencers: The History of Sequencing DNA. Genomics, 107(1), 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003
Heidenreich, M., & Zhang, F. (2016). Applications of CRISPR-Cas Systems in Neuroscience. Nature reviews. Neuroscience, 17(1), 36–44. https://doi.org/10.1038/nrn.2015.2
Herbst, S. M., Proepper, C. R., Geis, T., Borggraefe, I., Hahn, A., Debus, O., Haeussler, M., von Gersdorff, G., Kurlemann, G., Ensslen, M., Beaud, N., Budde, J., Gilbert, M., Heiming, R., Morgner, R., Philippi, H., Ross, S., Strobl-Wildemann, G., Muelleder, K., Vosschulte, P., … Hehr, U. (2016). LIS1-associated Classic Lissencephaly: A Retrospective, Multicenter Survey of the Epileptogenic Phenotype and Response to Antiepileptic Drugs. Brain & development, 38(4), 399–406. https://doi.org/10.1016/j.braindev.2015.10.001
Hopf, F. W., & Lesscher, H. M. (2014). Rodent Models for Compulsive Alcohol Intake. Alcohol (Fayetteville, N.Y.), 48(3), 253–264. https://doi.org/10.1016/j.alcohol.2014.03.001
Johnson, A. C., & Greenwood-Van Meerveld, B. (2016). The Pharmacology of Visceral Pain. Advances in pharmacology (San Diego, Calif.), 75, 273–301. https://doi.org/10.1016/bs.apha.2015.11.002
Kandel, E. R., Dudai, Y., & Mayford, M. R. (2014). The Molecular and Systems Biology of Memory. Cell, 157(1), 163–186. https://doi.org/10.1016/j.cell.2014.03.001
Lee, G. J., Park, J. H., & Park, H. K. (2008). Microdialysis Applications in Neuroscience. Neurological research, 30(7), 661–668. https://doi.org/10.1179/174313208X289570
Leroy, A., Foucher, J. R., Pins, D., Delmaire, C., Thomas, P., Roser, M. M., Lefebvre, S., Amad, A., Fovet, T., Jaafari, N., & Jardri, R. (2017). fMRI Capture of Auditory Hallucinations: Validation of the Two-Steps Method. Human brain mapping, 38(10), 4966–4979. https://doi.org/10.1002/hbm.23707
Liu, Z., Ding, L., & He, B. (2006). Integration of EEG/MEG with MRI and fMRI. IEEE engineering in medicine and biology magazine : the quarterly magazine of the Engineering in Medicine & Biology Society, 25(4), 46–53. https://doi.org/10.1109/memb.2006.1657787
Lodish H., Berk A., Zipurksy S. L. et. al., editors. (2000). Molecular Cell Biology, 4th edition. Freeman, p 94, 140, 147-148, 268-269.
Malik, A. N., Vierbuchen, T., Hemberg, M., Rubin, A. A., Ling, E., Couch, C. H., Stroud, H., Spiegel, I., Farh, K. K., Harmin, D. A., & Greenberg, M. E. (2014). Genome-wide Identification and Characterization of Functional Neuronal Activity-Dependent Enhancers. Nature neuroscience, 17(10), 1330–1339. https://doi.org/10.1038/nn.3808
Mayford, M., Siegelbaum, S. A., & Kandel, E. R. (2012). Synapses and Memory Storage. Cold Spring Harbor perspectives in biology, 4(6), a005751. https://doi.org/10.1101/cshperspect.a005751
Maze, I., Shen, L., Zhang, B., Garcia, B. A., Shao, N., Mitchell, A., Sun, H., Akbarian, S., Allis, C. D., & Nestler, E. J. (2014). Analytical Tools and Current Challenges in the Modern Era of Neuroepigenomics. Nature neuroscience, 17(11), 1476–1490. https://doi.org/10.1038/nn.3816
National Human Genome Research Institute. (July 2017). An Overview of the Human Genome Project. Accessed July 17, 2017 at https://www.genome.gov/12011238/an-overview-of-the-human-genome-project/
National Institute of Mental Health. (2017). Brain Stimulation Therapies. Accessed July 17, 2017 at https://www.nimh.nih.gov/health/topics/brain-stimulation-therapies/brain-stimulation-therapies.shtml
Olgiati, S., Quadri, M., & Bonifati, V. (2016). Genetics of Movement Disorders in the Next-Generation Sequencing Era. Movement disorders, 31(4), 458–470. https://doi.org/10.1002/mds.26521
Perry, R. H., Blessed, G., Perry, E. K., & Tomlinson, B. E. (1980). Histochemical Observations on Cholinesterase Activities in the Brains of Elderly Normal and Demented (Alzheimer-type) Patients. Age and ageing, 9(1), 9–16. https://doi.org/10.1093/ageing/9.1.9
Purves D, Augustine GJ, Fitzpatrick D, et al., editors. (2008). Neuroscience. 4th edition. Sinauer Associates, Inc. p 3-5, 16-17, 19-21, 25-27, 181-187, 465, 559, 673-674, 715-717.
Sejnowski, T. J., Koch, C., & Churchland, P. S. (1988). Computational Neuroscience. Science (New York, N.Y.), 241(4871), 1299–1306. https://doi.org/10.1126/science.3045969
Sokolowski M. B. (2001). Drosophila: Genetics Meets Behaviour. Nature reviews. Genetics, 2(11), 879–890. https://doi.org/10.1038/35098592
Svoboda, K., & Yasuda, R. (2006). Principles of Two-Photon Excitation Microscopy and its Applications to Neuroscience. Neuron, 50(6), 823–839. https://doi.org/10.1016/j.neuron.2006.05.019
Turek, F. W., Pinto, L. H., Vitaterna, M. H., Penev, P. D., Zee, P. C., & Takahashi, J. S. (1995). Pharmacological and Genetic Approaches for the Study of Circadian Rhythms in Mammals. Frontiers in neuroendocrinology, 16(3), 191–223. https://doi.org/10.1006/frne.1995.1007
US National Library of Medicine, National Institutes of Health. (2017). Genetics Home Reference – Huntington Disease. Accessed July 17, 2017 at https://ghr.nlm.nih.gov/condition/huntington-disease#genes
Usdin, K., & Kumari, D. (2015). Repeat-mediated Epigenetic Dysregulation of the FMR1 Gene in the Fragile X-related Disorders. Frontiers in genetics, 6, 192. https://doi.org/10.3389/fgene.2015.00192
Yoshino, K., Oka, N., Yamamoto, K., Takahashi, H., & Kato, T. (2013). Functional Brain Imaging Using Near-infrared Spectroscopy During Actual Driving on an Expressway. Frontiers in human neuroscience, 7, 882. https://doi.org/10.3389/fnhum.2013.00882
What to Read Next
Also In Tools & Techniques
Trending
Popular articles on BrainFacts.org